Mission
WHY DROSOPHILA GENITALIA?
- Genitalia evolve extremely quickly – we can observe many differences between species, even between the most closely related Drosophilids.
- The developing genitalia is easy to dissect and image when complex genital structures are forming.
- We can visualize individual cells in fixed tissues, which allows us to characterize the effects of genetic variation at high resolution.
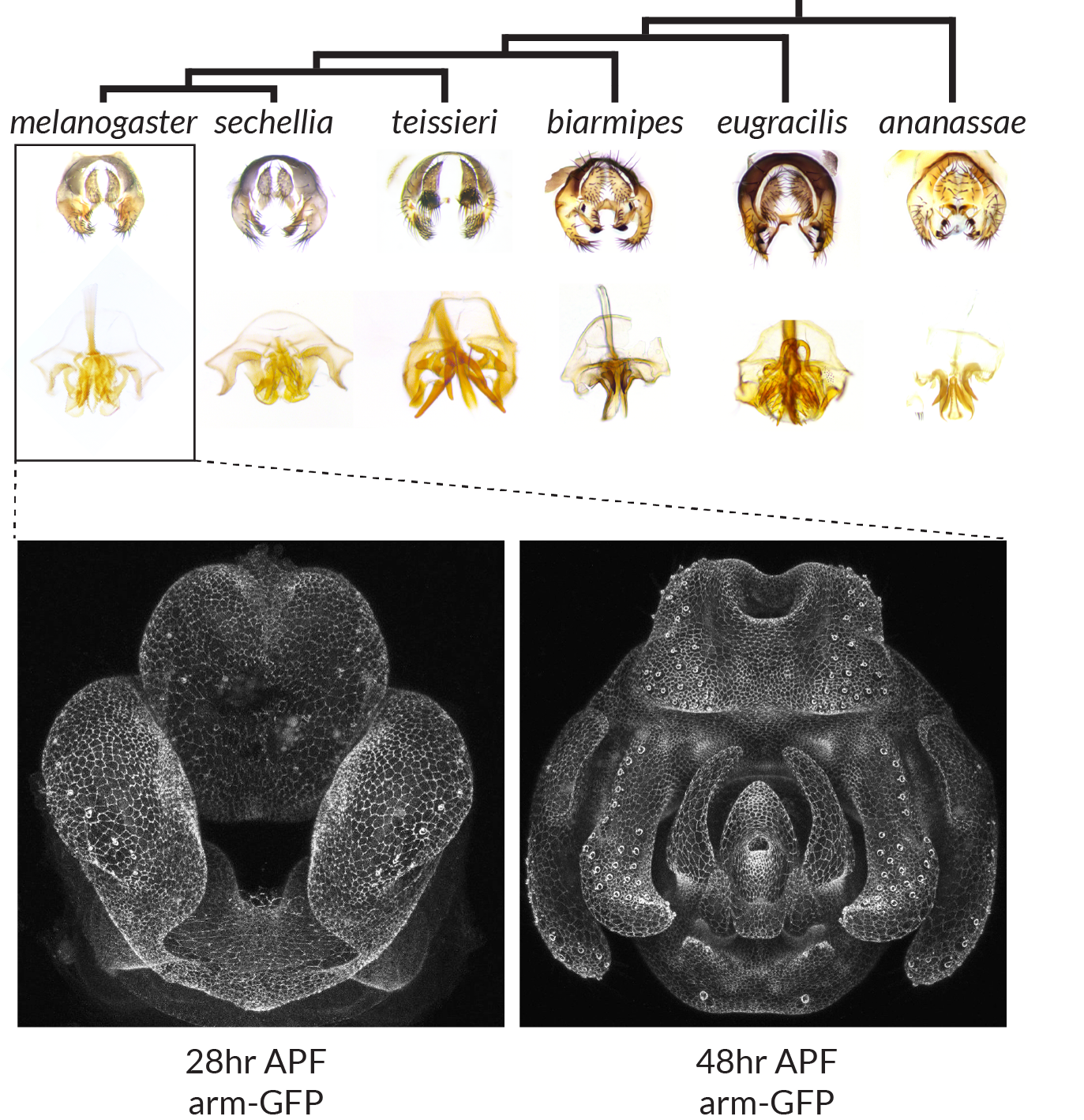
Evolution and development of Drosophila genitalia
Top: Adult morphology of the periphallus (top) and phallus (bottom) for different Drosophila species. Bottom: Confocal images of pupal genitalia at two time points during development from a transgenic Drosophila melanogaster line containing armadillo fused to GFP. APF: after puparium formation.
We’re inspired by foundational work in evolution and development, where important genetic changes often lie in regulatory regions that turn genes on and off in specific tissues [1–3]. However to understand how these mutations exert their effects, we need to know when and where genes are expressed in relevant tissues. For this type of data, we need only look to the Drosophila embryo, where decades of research has identified the transcription factors that form the body plan of the fly [4,5], and where their precise locations can be measured in exquisite quantitative detail [6–8]. Several groups have leveraged these resources to study body plan evolution [9–11], but body plans evolve slowly, which makes it difficult to identify causative mutations or characterize their effects.
To realize the potential of Drosophila genitalia as a model system, we’re building a map of transcription factors during pupal development.
This is when many genital structures form. We hope this resource will be useful to anyone interested in the emergence and divergence of these structures, and we are happy to apply our methods to additional genes suggested by the community.
OUR VALUES
Reproducibility
To build trust and ensure that other groups can reproduce our results, we have created detailed protocols on sample collection, probe design, in situ hybridization, imaging and analysis. For each gene, we also include details that vary between our experiments, such as our primers, probe concentration, probe amount, and color reaction length.
Transparency
In situ hybridization can be tricky, especially when done for a large number of genes. Results depend on probe quality, region of the gene, and may vary between samples. While we have curated representative images for each gene expression pattern, we also display multiple samples on each gene page to highlight the experimental variability.
We include annotations for the specific tissues where each transcription factor is expressed. These annotations are subjective and meant only as a useful guide; we encourage users to look at the raw images for any gene of interest, and to contact us if regarding questions or feedback on particular experiments.
Accessibility
In creating this resource, we want to support and expand the community of scientists interested in genital development and evolution. With this in mind, we designed our website to be easy to navigate and search. It is also dynamic – we will add new data as we collect it without going through a formal publication process. To understand the impact of genetic variation on genital morphology, we will need the collective effort of multiple groups. Given the complexity and diversity of genital morphology and the regulatory landscape as we understand it, there are many stories here, and room enough for many people to contribute.
OUR FUTURE
We started this venture by measuring the expression patterns of 100 highly expressed transcription factors in the genitalia. As we continue, we have three main goals, and we welcome feedback and collaboration from anyone interested in the future of this project.
Complete the Map
Our first goal is to complete measurements for all confirmed transcription factors expressed in the pupal genitalia. We will also measure expression patterns for other types of genes that may contribute to morphological changes, such as signalling molecules and cellular effectors. For genes that show strong patterns within the pupal genitalia, we hope to measure gene expression at cellular resolution with fluorescent in situ hybridization [12].
Expand the Map
We have currently focused on the male pupal genitalia of Drosophila melanogaster at two timepoints. Our second goal is to measure gene expression patterns in closely related species. As many structures have changed between species, we would predict some of these expression patterns may diverge. As these patterns are highly dynamic, we may need to incorporate additional timepoints into the atlas to identify cases of regulatory divergence.
While we have focused on male genitalia initially, we expect that many of the same transcription factors may specify genital structures in the female. We envision parallel atlases for both males and females in the future, which we hope will prove useful for anyone interested in the evolution of sexual dimorphism and sexual conflict.
Our database was designed with these future expansions in mind.
Interpret the Map
Transcription factors and signalling molecules are organized within developmental networks that dictate morphological outcomes. Now that we have established our initial map, our third goal is to determine how these genes regulate each other. We already find that many genes have reciprocal or overlapping expression patterns, and future work will follow up on specific hypotheses by perturbing genes with RNAi and measuring effects on gene expression and adult morphology.
We’re excited about the prospect of emerging single-cell sequencing technologies to rapidly generate gene expression/cell type atlases in complex tissues. We see the resource we’ve built as an important complement to these genomic datasets – spatial gene expression measurements are needed to identify cell types of interest and characterize how they change throughout development or in different species. We hope that the map will be useful in interpreting other types of genomic data, such as resolving quantitative trait loci to their causative genes [13–18].
REFERENCES
- Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. Nature Publishing Group; 2005;433: 481–487.
- Chan YF, Marks ME, Jones FC, Villarreal G Jr, Shapiro MD, Brady SD, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327: 302–305.
- Stern DL, Frankel N. The structure and evolution of cis-regulatory regions: the shavenbaby story. Philos Trans R Soc Lond B Biol Sci. 2013;368: 20130028.
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287: 795–801.
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, et al. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8: R145.
- Fowlkes CC, Hendriks CLL, Keränen SVE, Weber GH, Rübel O, Huang M-Y, et al. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell. 2008;133: 364–374.
- Garcia HG, Tikhonov M, Lin A, Gregor T. Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr Biol. 2013;23: 2140–2145.
- Gregor T, Garcia HG, Little SC. The embryo as a laboratory: quantifying transcription in Drosophila. Trends Genet. 2014;30: 364–375.
- Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. Functional evolution of a cis-regulatory module. PLoS Biol. 2005;3: e93.
- Wunderlich Z, Bragdon MD, Eckenrode KB, Lydiard-Martin T, Pearl-Waserman S, DePace AH. Dissecting sources of quantitative gene expression pattern divergence between Drosophila species. Mol Syst Biol. Nature Publishing Group; 2012;8: 604.
- Wotton KR, Jiménez-Guri E, Crombach A, Janssens H, Alcaine-Colet A, Lemke S, et al. Quantitative system drift compensates for altered maternal inputs to the gap gene network of the scuttle fly Megaselia abdita. Elife. 2015;4. doi:10.7554/eLife.04785
- Luengo Hendriks CL, Keränen SVE, Fowlkes CC, Simirenko L, Weber GH, DePace AH, et al. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution I: data acquisition pipeline. Genome Biol. BioMed Central Ltd; 2006;7: R123.
- Macdonald SJ, Goldstein DB. A quantitative genetic analysis of male sexual traits distinguishing the sibling species Drosophila simulans and D. sechellia. Genetics. 1999;153: 1683–1699.
- Zeng ZB, Liu J, Stam LF, Kao CH, Mercer JM, Laurie CC. Genetic architecture of a morphological shape difference between two Drosophila species. Genetics. 2000;154: 299–310.
- Masly JP, Dalton JE, Srivastava S, Chen L, Arbeitman MN. The genetic basis of rapidly evolving male genital morphology in Drosophila. Genetics. 2011;189: 357–374.
- McNeil CL, Bain CL, Macdonald SJ. Multiple Quantitative Trait Loci Influence the Shape of a Male-Specific Genital Structure in Drosophila melanogaster. G3 . 2011;1: 343–351.
- Tanaka KM, Hopfen C, Herbert MR, Schlötterer C, Stern DL, Masly JP, et al. Genetic Architecture and Functional Characterization of Genes Underlying the Rapid Diversification of Male External Genitalia Between Drosophila simulans and Drosophila mauritiana. Genetics. Genetics Society of America; 2015;200: 357–369.
- Takahara B, Takahashi KH. Genome-Wide Association Study on Male Genital Shape and Size in Drosophila melanogaster. PLoS One. 2015;10: e0132846.